What Scientists Say
Our roots

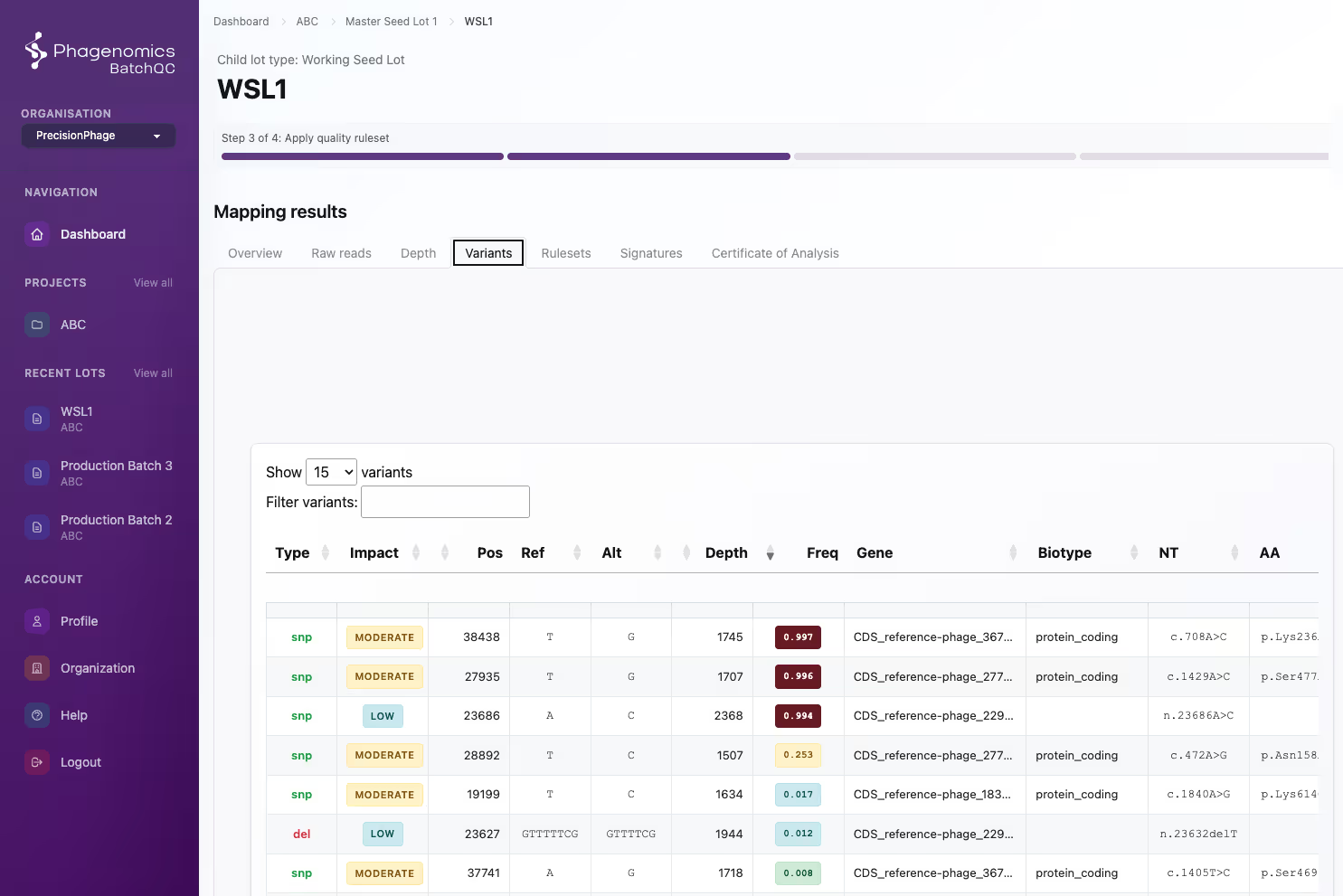
We Are Serious About Our Science
Developed by phage researchers with more than 40 years of combined experience in phage research, integrating deep expertise in modern bioinformatics and computational analysis.
Phagenomics software leverage Finland’s supercomputing infrastructure to enable large-scale and high-speed bioinformatics analyses.
We are committed to upholding the highest bioinformatics standards and continually developing Phagenomics software to meet the evolving requirements of science and industry.
Phagenomics Pricing
Flexible plans for people pushing the boundaries of genomic science.



Combining rich academic background with state-of-the-art computational solutions
Phagenomics is developed by PrecisionPhage, a company dedicated to furthering groundbreaking research and innovative solutions that drive success in biotechnology. Our commitment to excellence has established us as a trusted partner in the industry.
Years of combined phage research experience
Phage genomes assembled
Phage genomes annotated
of academic papers
FAQs
If your question is not listed below, don’t hesitate to contact us!
Yes. We have invested considerable effort in providing you with a clean and intuitive graphical user interface. There is no need for coding or command lines.
Phagenomics Core automates phage genome assembly and annotation for research and discovery. BatchQC extends this to GMP-level quality control—verifying genome integrity, detecting mutations, and generating regulatory-ready reports for pharmaceutical-grade phage production.
Our team combines over 40 years of academic and applied phage research experience from leading Finnish universities and global collaborations. Founders and key experts hold PhDs in microbiology, phage therapy, and bioinformatics, bridging both laboratory and computational science.
Yes. Our Finland-based laboratory offers phage isolation, characterization, DNA preparation, sequencing, and R&D services for research and preclinical development. We do not provide direct patient treatments, but we support clinicians and companies developing therapeutic phages.
Accelerate Your
Phage Bioinformatics Today